The following code demonstrates the use of unfold() with
artificial data.
We first create some artificial data of which we know the correct
configuration. a1 and a2 are the coordinates
of a first group of points, which form a circle (with some very small
perturbations added). b1 and b2 are the
coordinates of a second group of points, which form a square.
r <- seq(from=0,to=2*pi,length=24)
a1 <- cos(r)*4 + 0.00001*rnorm(r)
a2 <- sin(r)*4 + 0.00001*rnorm(r)
b1 <- c(.5,-.5,-.5,.5)*3 + 5
b2 <- c(.5,.5,-.5,-.5)*3 + 1Im the next step, we compute the squred distamces between the two groups of points.
Now we call unfold():
2-dimensional unfolding solution
Group 1: 24 points
Group 2: 4 points
Stress: 2.709978e-23The printout indicates that unfold() prefers a
two-dimensional solution, which fits the observed squared distances
perfectly.
The output does not tell us, who well we can recreate the original configuration, so let’s do some plotting.
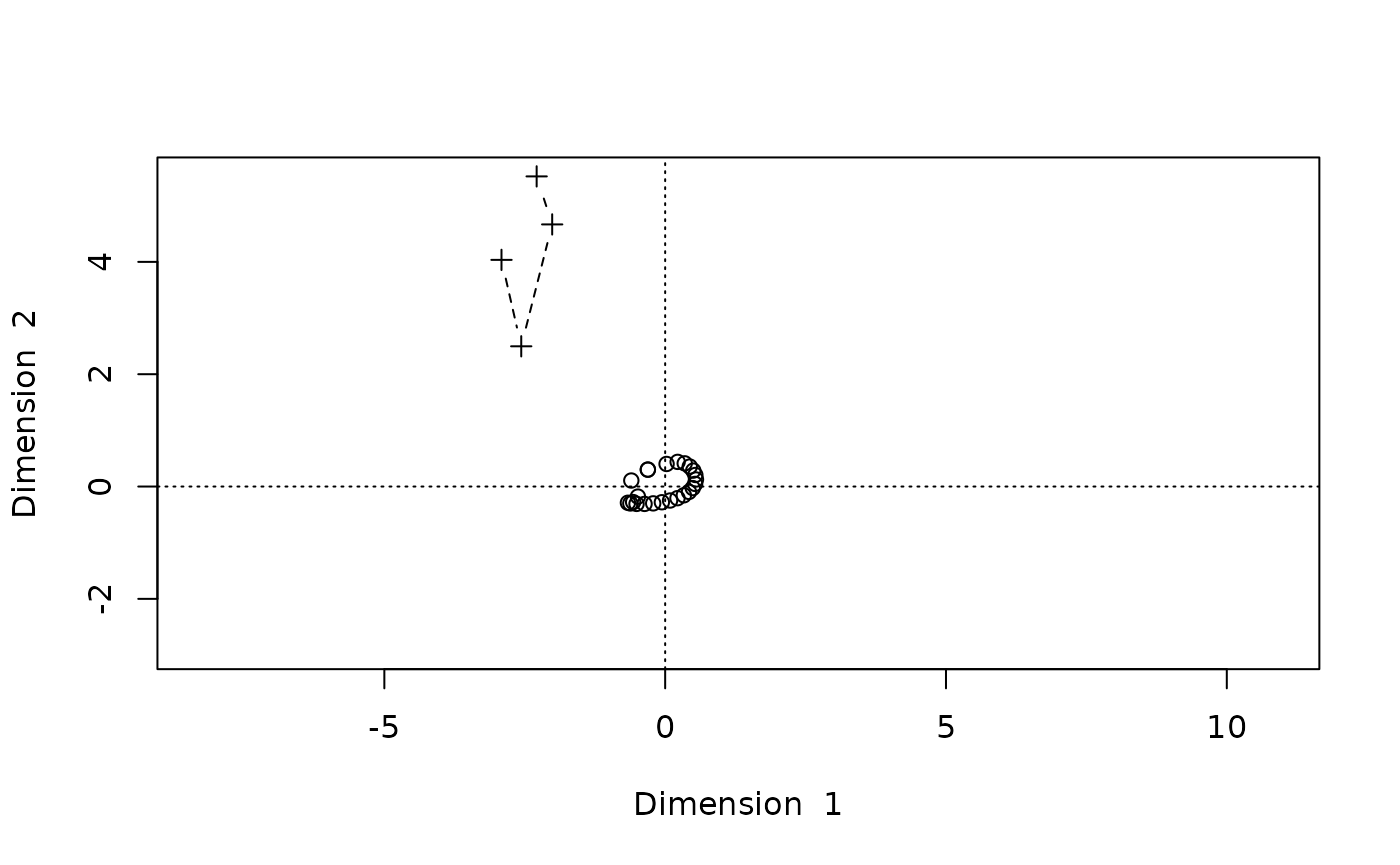
With biplot() we get an illustration of the
reconstructed configuration. Second, let’s plot the reconstructed
locations of the two groups of points. To enhance the geometric
configuration of the points, we require a points-and-line plot by using
the argument type="b"
biplot(Dsq.uf,type="b")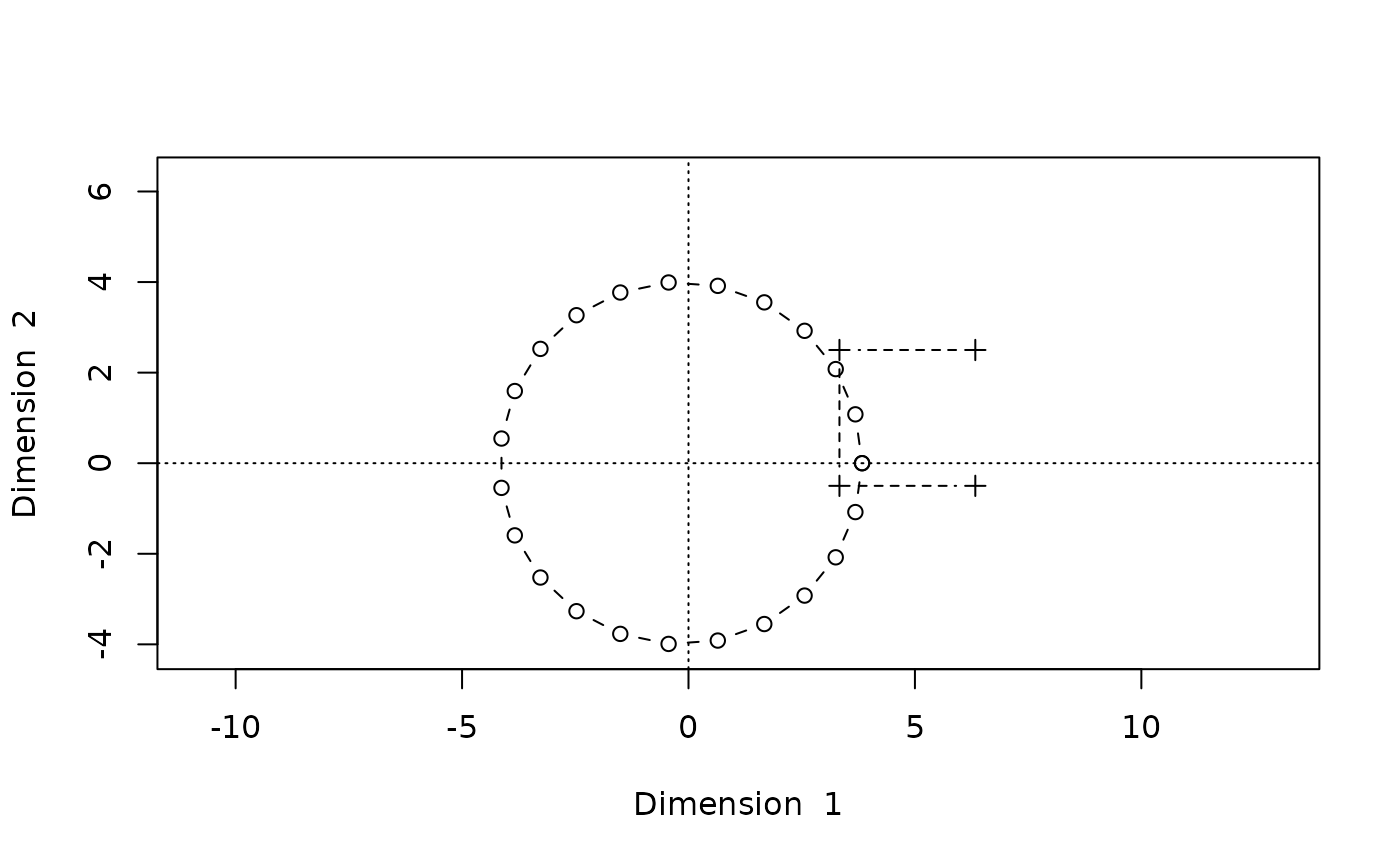
Now let’s compare this with the original configuration, where we need to make sure that all of the original points appear in the plot.
A <- cbind(a1,a2)
B <- cbind(b1,b2)
orig <- rbind(A,B)
xlim <- ylim <- range(orig)#*1.5
plot(A,type="b",pch=1,asp=1,
xlim=xlim,ylim=ylim,
xlab="Dimension 1",ylab="Dimension 2")
lines(B,type="b",pch=3,lty=2)
abline(h=0,v=0,lty=3)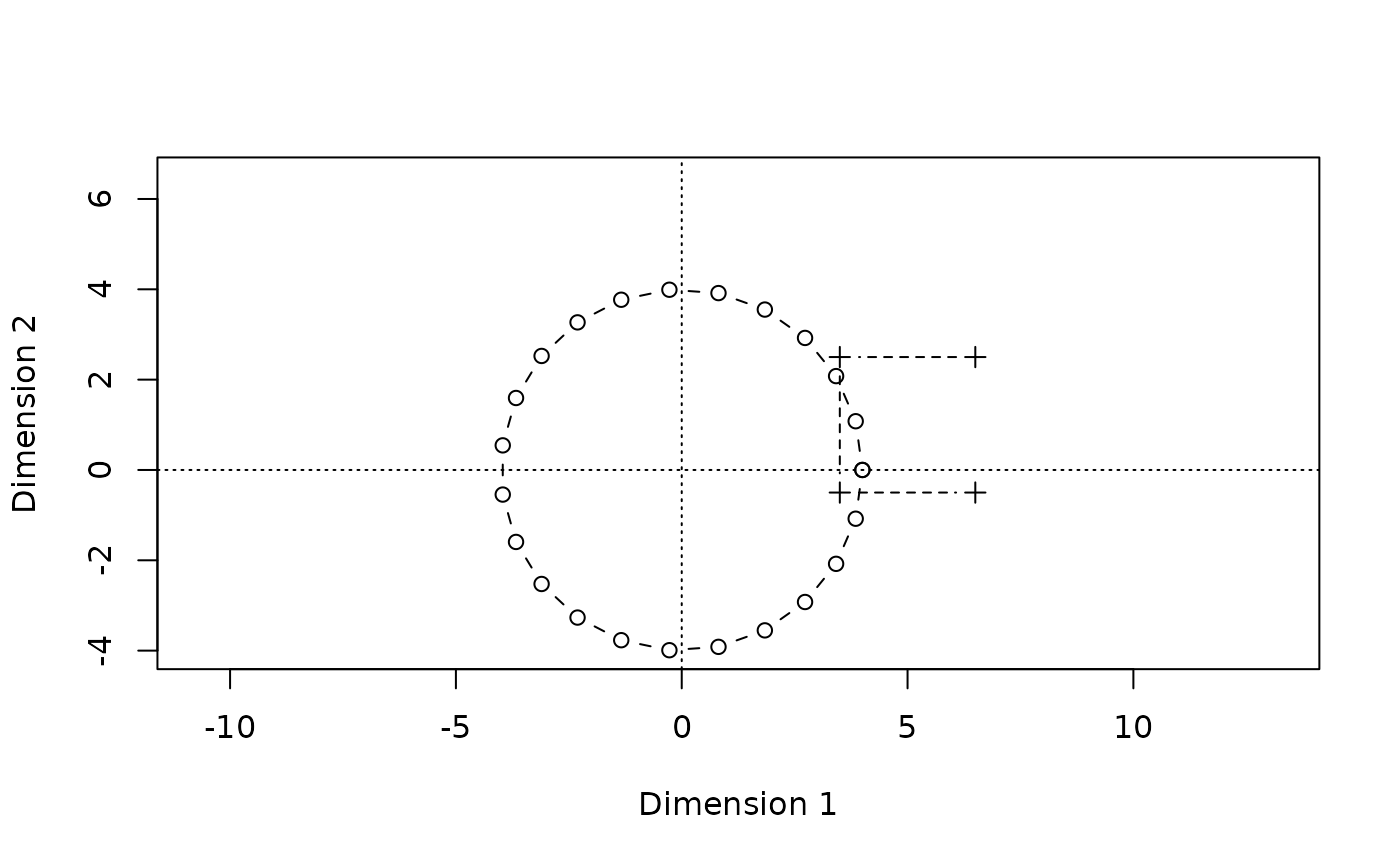
This looks pretty similar, but a comparison will be easier if we put both plots side by side:
oldpar <- par(mfrow=c(1,2))
plot(A,type="b",pch=1,
xlim=xlim,ylim=ylim,
xlab="Dimension 1",ylab="Dimension 2",main=expression("Original data"),asp=1)
lines(B,type="b",pch=3,lty=2)
abline(h=0,v=0,lty=3)
biplot(Dsq.uf,type="b",
xlim=xlim,ylim=ylim,
main=expression(paste(italic(unfold)," solution")),asp=1)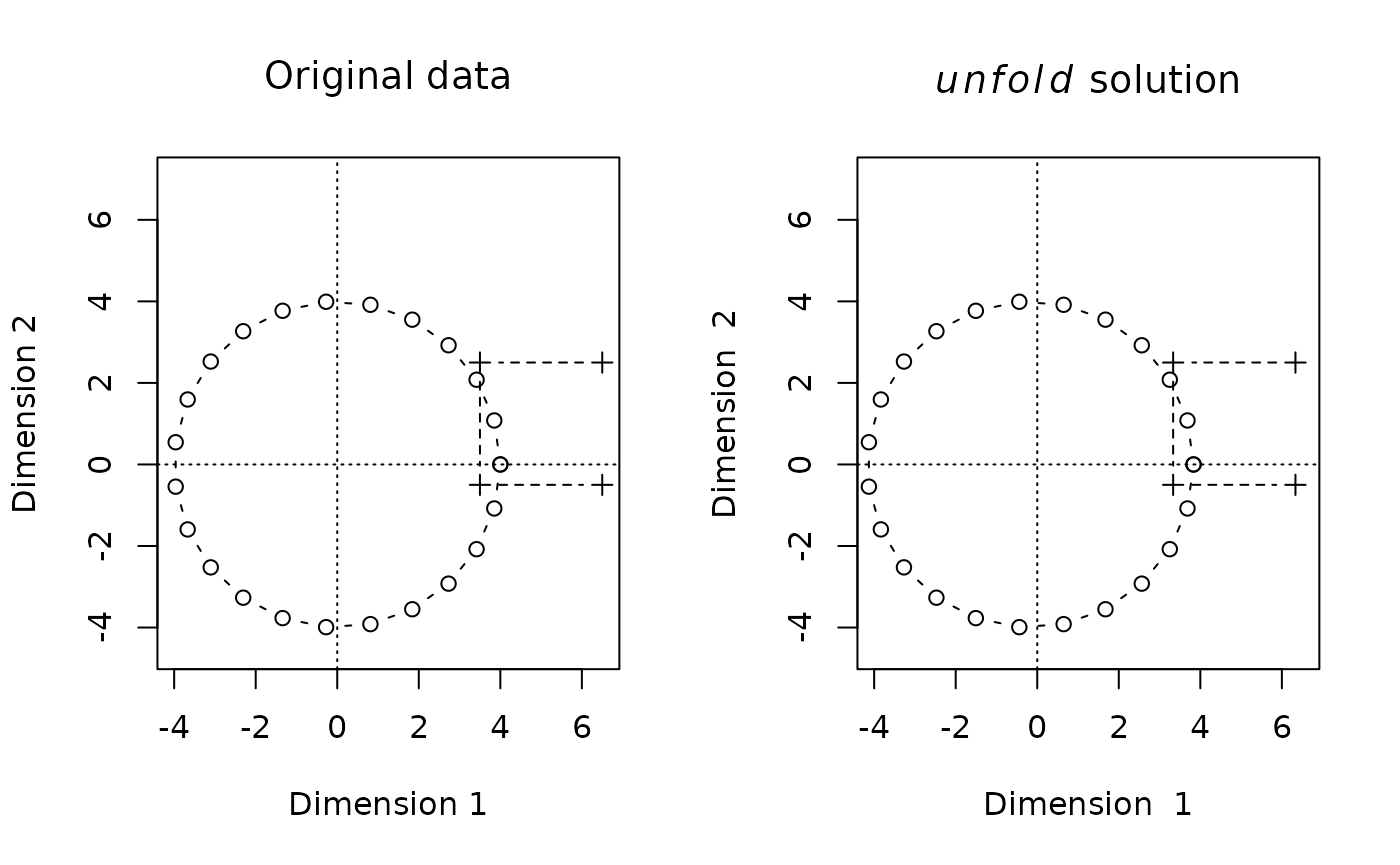
par(oldpar)Curiously, Schönemann’s algorithm fails with a perfect circle:
r <- seq(from=0,to=2*pi,length=24)
a1_0 <- cos(r)*4 # NB: no random noise added here
a2_0 <- sin(r)*4
b1_0 <- c(.5,-.5,-.5,.5)*3 + 5
b2_0 <- c(.5,.5,-.5,-.5)*3 + 1
D1_0 <- outer(b1_0,a1_0,"-")
D2_0 <- outer(b2_0,a2_0,"-")
Dsq0 <- D1_0^2+D2_0^2
Dsq.uf0 <- unfold(Dsq0,squared=TRUE)
Dsq.uf0
2-dimensional unfolding solution
Group 1: 24 points
Group 2: 4 points
Stress: 0.2191948The stress level indicates an imperfect fit, while the configuration within groups seems to be okay, the overall configuration is wrong:
biplot(Dsq.uf0,type="b")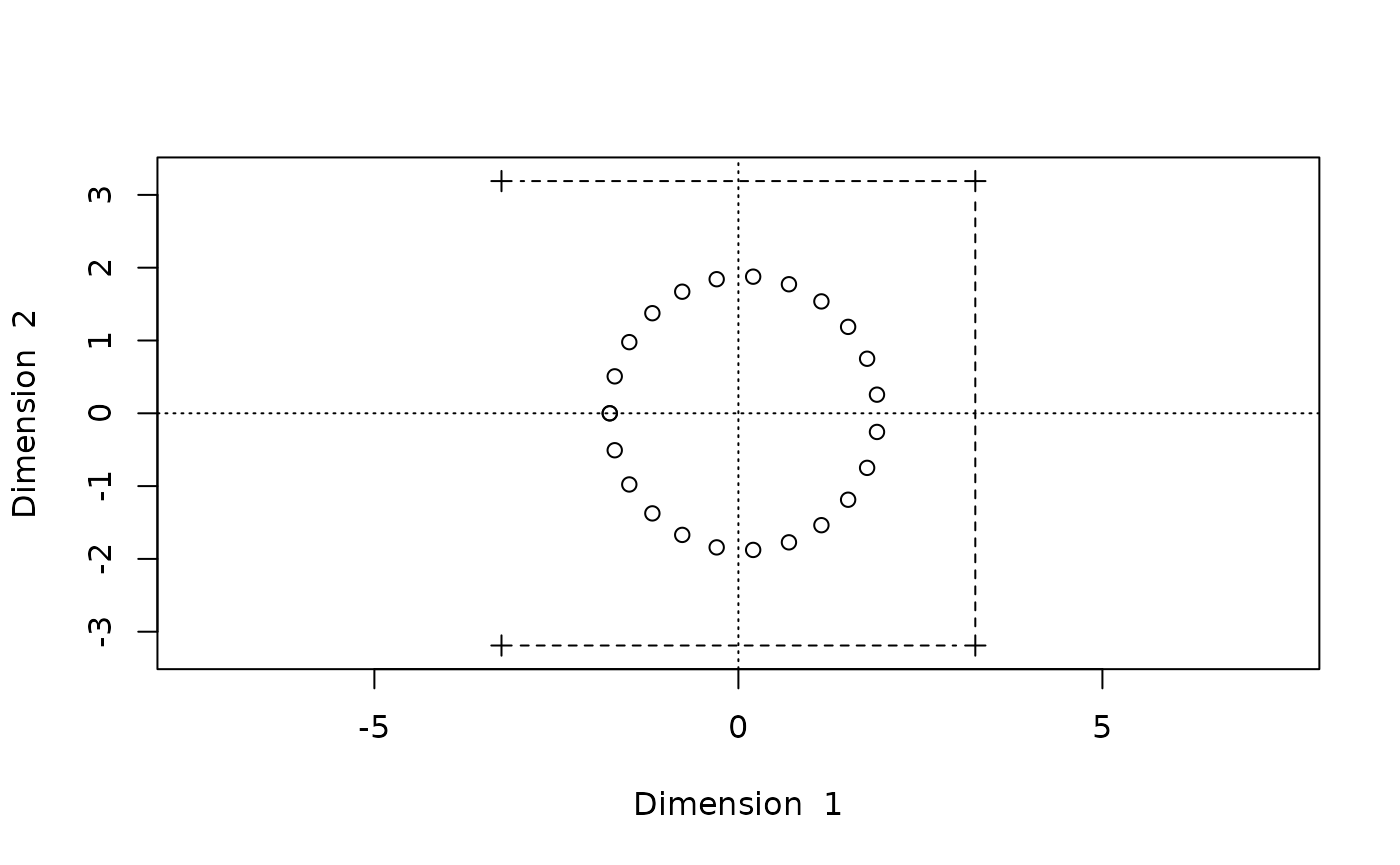
However, the iterative conjugate gradient method seems to work:
Dsq.uf0.cg<-unfold(Dsq,squared=TRUE,method="CG")
Stress for starting values 3.774471e-23
Stress after translation 3.774471e-23
Stress after rescaling 4.468749e-24
Gradient: -2.769453e-07 3.325053e-07
Stress after rigid transformation 4.468749e-24
Gradient: -2.769453e-07 3.325053e-07 -7.07713e-08 3.910066e-08
Dsq.uf0.cg
2-dimensional unfolding solution
Group 1: 24 points
Group 2: 4 points
Stress: 4.141668e-24We get a non-convergence warning, but the stress level is much lower and the reconstructed configuration looks okay:
biplot(Dsq.uf0.cg,type="b")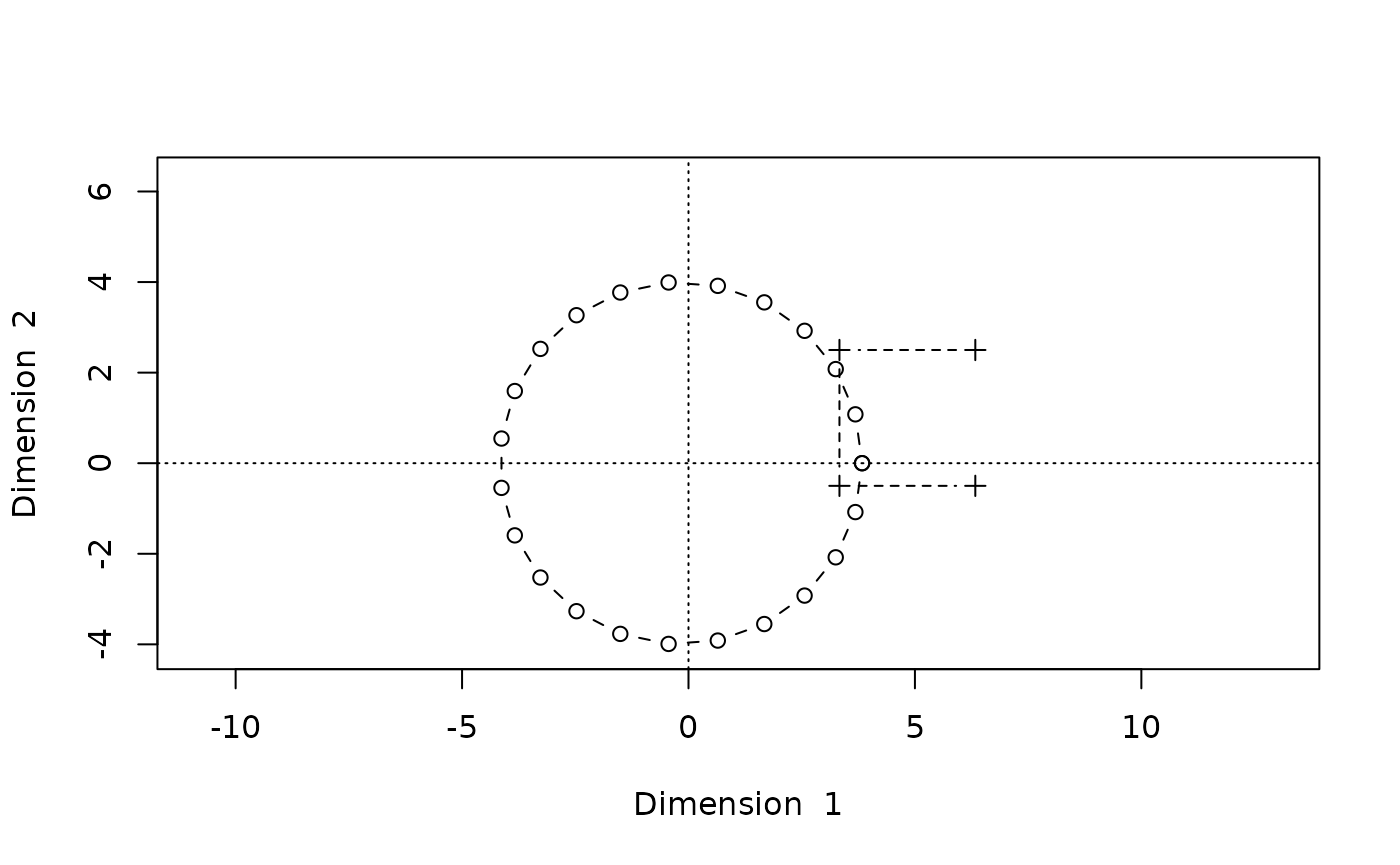
Less surprisingly, it is important to correctly indicate whether the
input for unfold() consists of squared distances or
un-squared distances.
Here we provide squared distances, but indicate that the distances are not squared.
Dsq.uf1 <- unfold(Dsq,squared=FALSE)
Dsq.uf1
3-dimensional unfolding solution
Group 1: 24 points
Group 2: 4 points
Stress: 8861670We get an additional spurious dimension and the reconstruction of the points is distorted:
biplot(Dsq.uf1,type="b")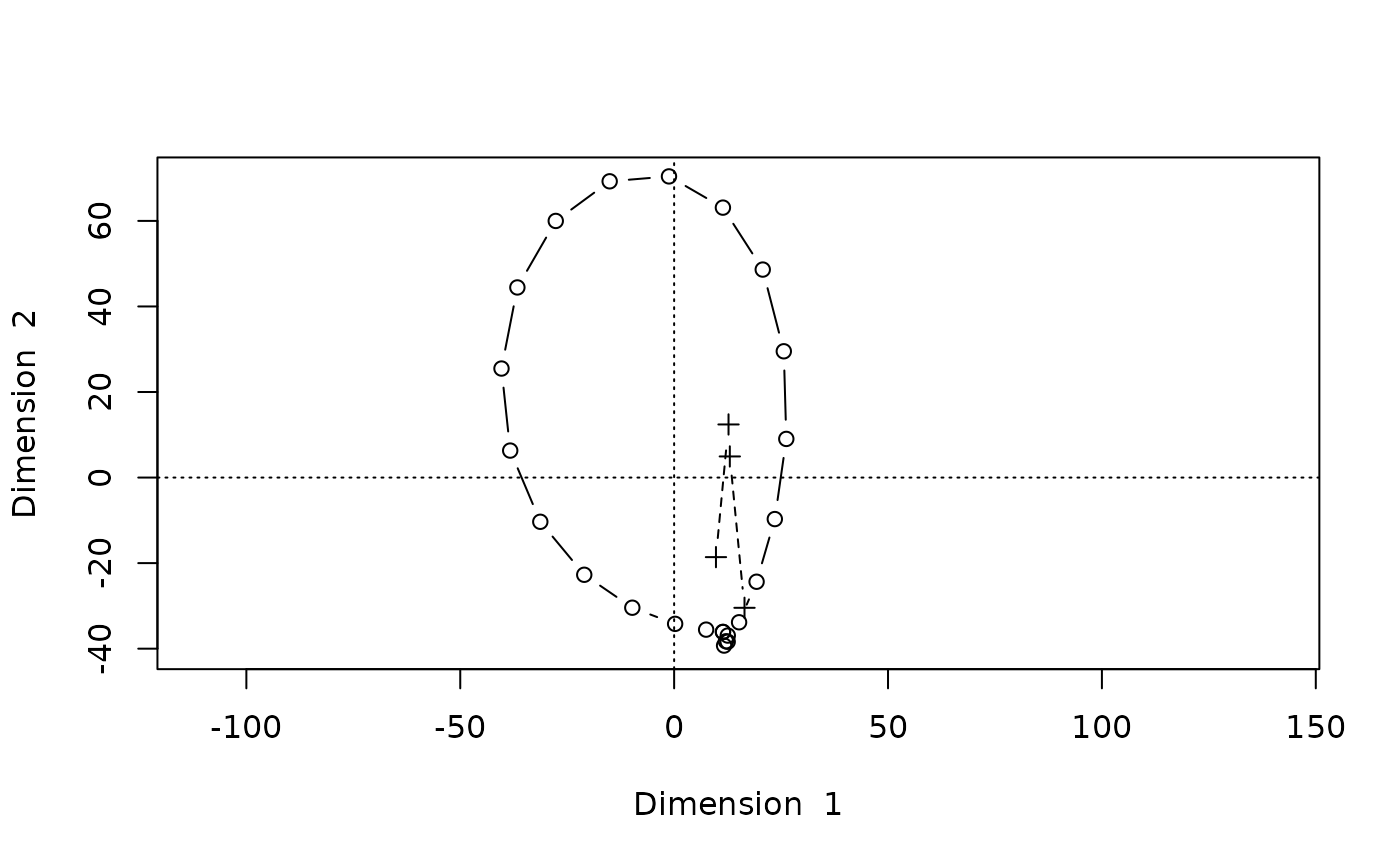
Here we provide non-squared distances, but indicate that the distances are squared.
3-dimensional unfolding solution
Group 1: 24 points
Group 2: 4 points
Stress: 799782.4Again we end up with a spurious additional dimension and a distorted configuration of the points:
biplot(Dsq.uf2,type="b")